7 Restriction enzymes
Learning Objectives
-
To define restriction enzymes and describe their biological role in prokaryotic defense systems.
-
To predict the cutting pattern of a restriction enzyme on a given DNA sequence, including identification of recognition sites and resulting fragment sizes.
-
To design and interpret a restriction digest experiment, including how to select enzymes and analyze fragment patterns using gel electrophoresis.
What are restriction enzymes?
Restriction enzymes, or restriction endonucleases, are proteins produced by bacteria to defend against foreign DNA by cutting it into nonfunctional pieces. These enzymes act as DNA scissors, specifically recognizing and cleaving DNA at restriction sites. A nuclease is any enzyme that cleaves the DNA backbone by breaking phosphodiester bonds. Endonucleases cut within a DNA molecule at specific sequences, while exonucleases cut from a free DNA end. Exonucleases typically act on single-stranded DNA (ssDNA) or nicks in double-stranded DNA (dsDNA), whereas endonucleases act exclusively on intact dsDNA.
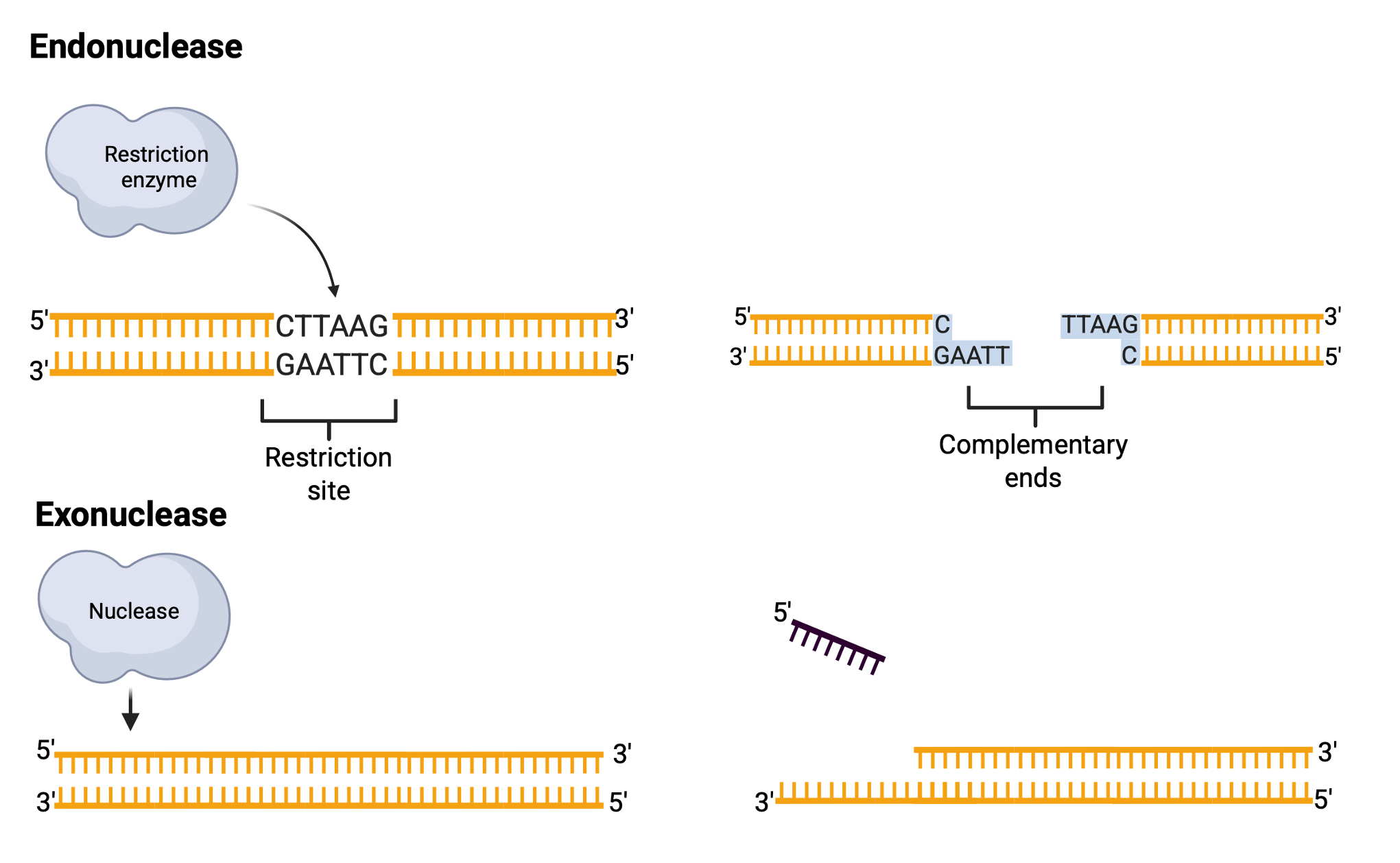
Restriction sites are specific 4- or 6-base-pair palindromic sequences, where the 5′-to-3′ sequence on one strand matches the 5′-to-3′ sequence on the complementary strand. For instance, the sequence 5′ GAATTC 3′ is a palindrome and serves as the restriction site for EcoRI, a restriction enzyme derived from Escherichia coli RY13.

EcoRI cuts the DNA between the G and A in each strand, creating single-stranded “sticky ends” that can easily rejoin with complementary sticky ends. Not all restriction enzymes produce sticky ends; some, like SmaI, generate “blunt ends” by cutting both strands at the same position. Restriction enzymes widely used in laboratories typically recognize specific palindromic sequences and cleave the DNA at precise positions within these sites. The cleavage pattern of restriction enzymes results in DNA fragments whose sizes and numbers depend on the locations of restriction sites within the DNA molecule. For instance, sequences of 4 bases occur randomly approximately once every few hundred bases, while 6-base sequences occur less frequently.
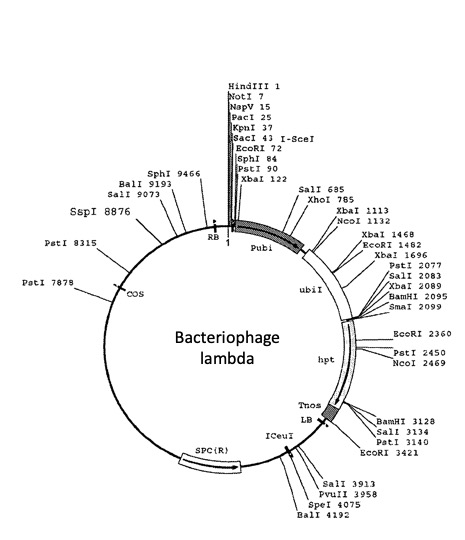
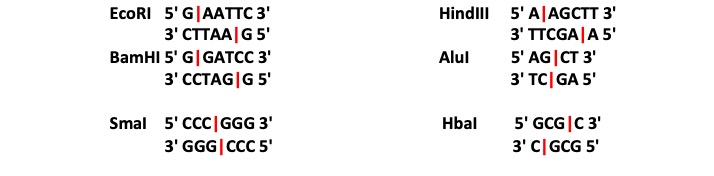 Under suitable conditions of salt concentration, pH, and temperature, a restriction enzyme can produce consistent DNA fragment patterns. Some DNA molecules, such as bacteriophage lambda (~48,000 base pairs), have diverse restriction sites, while others may lack specific restriction sites altogether. Restriction enzymes are indispensable tools in molecular biology for DNA analysis, cloning, and other applications.
Under suitable conditions of salt concentration, pH, and temperature, a restriction enzyme can produce consistent DNA fragment patterns. Some DNA molecules, such as bacteriophage lambda (~48,000 base pairs), have diverse restriction sites, while others may lack specific restriction sites altogether. Restriction enzymes are indispensable tools in molecular biology for DNA analysis, cloning, and other applications.
What are some applications for restriction enzymes?
Restriction enzymes (restriction endonucleases) are proteins that recognize specific DNA sequences and cleave the phosphodiester backbone at or near those sites. Their discovery provided a reliable way to cut DNA into defined, reproducible fragments, enabling modern genetic engineering techniques.
Before restriction enzymes were available, adding foreign DNA to a host cell was not possible because:
Chromosome size: Bacterial chromosomes are large molecules containing thousands of genes. Eventhough genes could be identified functionally (e.g., lactose fermentation) and mapped genetically to nearby markers, genes could not be physically located, isolated, or manipulated.
Fragmentation methods: DNA could only be broken into smaller pieces by physical shearing, producing random fragments. Random fragments were not reproducible between preparations.
Cloning limitations: Simple DNA fragments are degraded rapidly in most bacteria due to nucleases. Cloning requires vectors (e.g., plasmids, viruses), which must be cut at specific sites to insert new DNA and without precise cutting, insertion was not possible.
With restriction enzymes:
Defined cutting: Cleavage at specific recognition sites yields the same fragment pattern every time, so defined DNA fragments can be ligated into vectors to create new DNA molecules (recombinant DNA) from multiple sources.

Let’s cut a viral genome with restriction enzymes, on paper: In silico lambda DNA digestion (adapted from The American Phytopathological Society APS).
In silico = in or on a computer, done or produced by using computer software or simulation. In other words, you’ll practice on paper an enzymatic restriction digestion before you do it on DNA.
- Download and SAVE the Lambda DNA sequence (lambdafasta).
- Save the file on your computer.
- Open the file in Microsoft Word. It contains the entire phage Lambda DNA genome.
- Determine how many letters there are in the genome. This number correspond to the number of bases of genome size in bp.
Simulating the Effects of Restriction Enzymes
Remember that there are a large number of restriction endonucleases (restriction enzymes), and that each enzyme recognizes a specific sequence of DNA nucleotides and cuts at a specific point within that sequence. The four restriction enzymes we will use for this exercise are BamHI, EcoRI, HindIII and AluI.
For each enzyme perform a “single digest” simulation and count the number and size of fragments produced using the following procedure:
- Position the cursor on the first letter of your sequence (the first line starting with the symbol “>” is the name of the sequence).
- Using the SEARCH option available on your operating system look for the motif that correspond to each enzyme’s restriction site (e.g. GAATTC). The number of hits corresponds to the number of restriction sites within your complete sequence.
- Position the cursor after the first letter of the restriction site and select the text “previous” to it. Using the word count option of your text editor obtain the number of characters from the beginning of the sequence. This number will give you the first position of the restriction site and the size of the fragment.
- Repeat for each restriction enzyme.
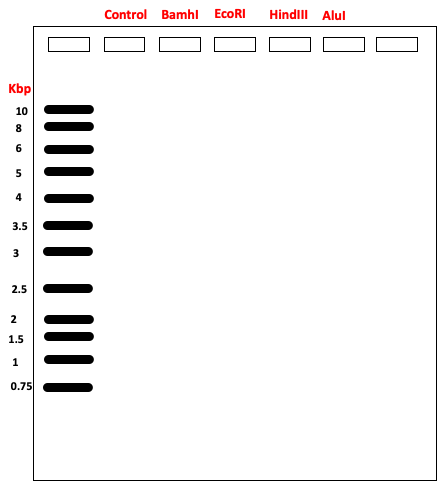
- Indicate the expected fragment sizes on the following “agarose gel”. Use the DNA ladder lane as a guide.
Materials
- Four Eppendorf microtubes
- Microtube rack
- Micropipette set
- Beaker or foam cup with crushed ice for the following
- 20 µl of 0.4 µg/µl λ DNA
- 5 µl BamHI restriction enzyme
- 5 µl EcoRI restriction enzyme
- o 5 µl HindIII restriction enzyme
- Restriction enzymes buffers
- 10 µl distilled water
- Electrophoresis chamber
- Power supply
- 20 µl 10X loading buffer
- 1.0% agarose gel
Safety concerns
This experiment uses chemicals, biological materials, and equipment that pose some potential risks:
- Handle the ethanol and the kit reagents with gloves, at all times.
- Use biosafety protocols to handle the DNA samples to prevent accidental exposure, and dispose of all the waste in a biohazard container.
- Make sure you balance the microcentrifuges correctly before you use them.
- Avoid eating or drinking, and manage spills and emergencies following the directions of the instructor.
Protocol
- Put on gloves. Keep all enzymes and DNA aliquots on ice through step 6.
- Label 2 PCR tubes with the name of the enzymes indicated below, and place them in the tube rack.
- Spin down the tubes before opening them to ensure the liquid is collected at the bottom.
- Add each reagent in the order indicated, making sure to transfer the complete volume.
- Follow the directions given by your instructor.
- Close the microtubes, mix the content by flicking the tubes, spin the tubes down and place them on ice.
- Incubate the tubes following your instructor’s directions.
- Freeze the digested samples or run on a 1% agarose gel along a 1Kb DNA ladder.
Key Takeaways
-
Restriction enzymes recognize and cut DNA at specific sequences, usually palindromic, making them essential tools for DNA manipulation in cloning, mapping, and analysis.
-
Different enzymes produce different types of ends—blunt or sticky (cohesive)—which influence how DNA fragments can be joined or recombined.
-
Restriction digests are highly predictable and reproducible, allowing researchers to plan experiments with precision and analyze DNA fragment patterns through gel electrophoresis.
Lecture slides
Download the slides to follow the pre-lab lecture. enyzmaticdigestion_2025
Bacteriophage lambda genome
Download Word file for the in-silico analysis: lambdafasta
Lab report
Download the EnzymaticRestrictionLabReport file and submit on Canvas, following the directions of your instructor.
