5 RNA extraction and purification
Learning Objectives
- To recognize the role of reagents (e.g., detergents, salts, enzymes) and understand their function in the isolation of RNA.
- To recognize the activity of RNAses and how they affect RNA quality.
- To learn to control RNAses activity and minimize RNA degradation.
- To visualize and confirm RNA extraction using agarose electrophoresis and learn to differentiate it from DNA.
- To discuss how extracted RNA can be used in downstream applications, such as cDNA synthesis and quantitative PCR.
What is the purpose of extracting RNA from cells?
The main purpose to extracting high-quality RNA is to study the expression of genes in cells or tissues, allowing researchers to measure transcript levels (or gene copy number) and to understand how cells respond to environmental stimuli or stress, such as exposure to space or space-like conditions, or any other type of treatments that changes the physiology of the cell. It is also a fundamental step for transcriptomic studies, such as RNA sequencing (RNA-seq), which profiles mRNA, non-coding RNAs, and small RNAs to explore gene regulation at the whole-genome level. RNA can be used to detect RNA viruses (e.g., SARS-CoV-2) and to monitor the expression of biomarkers for diseases such as cancer. Similarly, it has allowed the development of RNA-based therapies, including mRNA vaccines and RNA interference (RNAi) technologies used to treat a wide range of diseases. Beyond medicine, RNA extraction facilitates space, agricultural and environmental research by studying gene expression in plants and microbial communities, aiding in crop improvement and ecosystem monitoring.
What are the differences between RNA and DNA extraction methods?
| RNA Extraction | DNA Extraction | |
| Molecules extracted | mRNA, tRNA, rRNA, and non-coding RNAs | Nuclear and mitochondrial DNA |
| Stability | Highly unstable and easily degraded by RNAses activity | More stable and resistant to environmental degradation than RNA |
| Enzymatic Protection | Requires strict precautions o prevent RNases contamination, including RNase-free reagents and tools (DEPC-treated) | Relatively resistant to DNases activity, particularly in the presence of Mg+2 ions chelators such as EDTA |
| Lysis Buffer | Often includes reducing agents such as guanidine isothiocyanate or beta-mercaptoethanol to inactive RNases. | Typically contains EDTA, detergents and salts to lyse cells and stabilize DNA |
| Purification | Usually involves phenol-chloroform extraction or silica column purification specific for RNA. | It uses methods similar to those for RNA, but the buffers and binding conditions are optimized for DNA |
| Precipitation | Requires isopropanol or ethanol for RNA precipitation, often with co-precipitants like glycogen | Precipitation protocols are similar, typically using ethanol or isopropanol |
| Quality | Spectrophotometry (A260/A280 and A260/A230 ratios) and agarose or capillary (Bioanalyzer or Tapestation) electrophoresis | Similar methods, but DNA integrity is assessed based on high-molecular-weight bands |
| Storage | Stored at –80°C in RNase-free water or buffer; avoid freeze-thaw cycles to prevent degradation | Stored at –20°C or –80°C in TE buffer or water for long-term stability |
| Applications | Studying gene expression (e.g., qPCR, RNA-seq); Functional studies of non-coding RNAs; Viral RNA detection | Genotyping, sequencing, and cloning; Genome studies and mutation analysis; DNA barcoding |
What are RNases and how can we control their activity?
RNases are enzymes (proteins) that break the phosphodiester bonds in RNA, breaking it into random-size fragments. They are resistant to many methods of decontamination, including autoclave temperatures, and require strong reducing chemicals to remove them from surfaces and solutions.
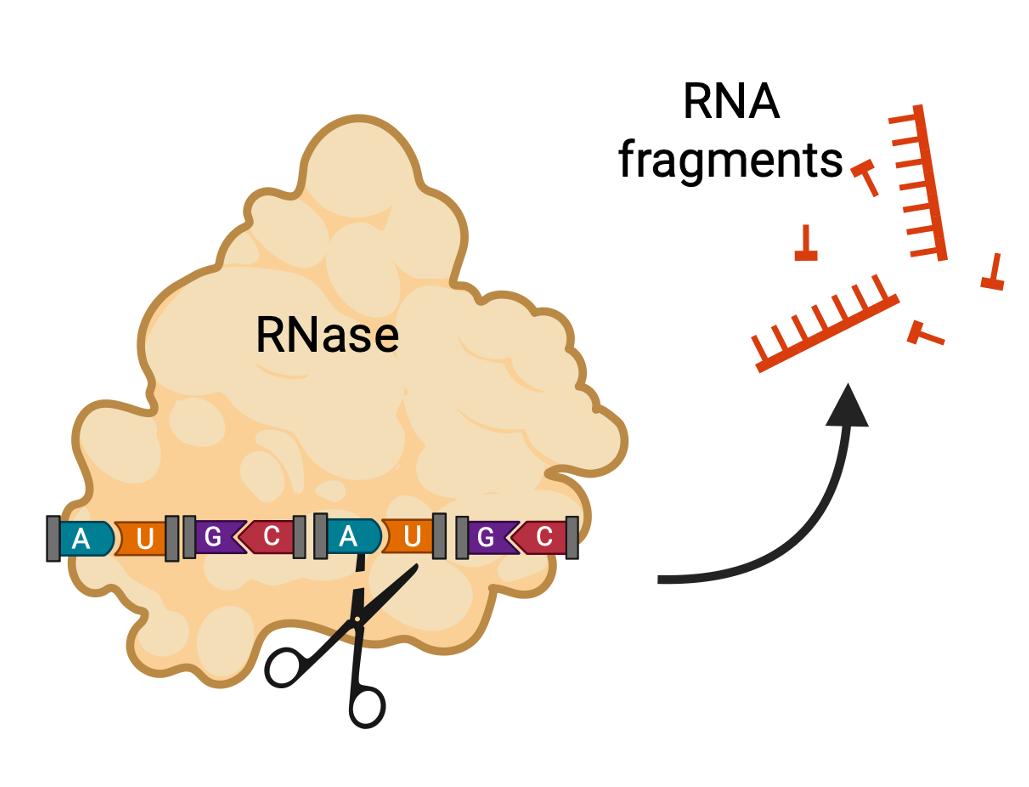
To protect from RNases in lab, RNA samples must be properly stored, buffers and solutions are tested for RNase contamination, and working spaces and equipment must be regularly treated with RNase inhibitors (e.g. RNAZap).
The presence of just trace amounts of RNase compromises RNA integrity even if the samples are stored frozen. For short-term storage, RNA samples can be resuspended in RNase-free water (with 0.1 mM EDTA) or TE buffer (10 mM Tris, 1 mM EDTA) and stored at –80°C. For long-term storage, perform a salt/alcohol precipitation and store it at –20°C. The low temperature and the presence of alcohol inhibit all enzymatic activity. A slighlty acidic pH (due to the presence of sodium acetate or ammonium acetate) also helps stabilize the RNA. Note that the RNA will have to be centrifuged out of this solution prior to any downstream application.
DNA/RNA Shield (Zymo Research)
DNA/RNA Shield is a DNA and RNA transport and storage medium for any biological sample. DNA/RNA Shield preserves the genetic integrity and expression profiles of samples at ambient temperatures (no refrigeration or freezing needed) and completely inactivates infectious agents (viruses, bacteria, fungi, & parasites). Nucleic acids from samples stored in this reagent can be isolated directly without precipitation or reagent removal (compatible with most DNA and RNA purification kits).
Trizol (Invitrogen)
TRIzol® Reagent allows you to perform sequential precipitation of RNA, DNA, and proteins from a single sample. After homogenizing the sample with TRIzol® Reagent, chloroform is added, and the homogenate is allowed to separate into a clear upper aqueous layer (containing RNA), and interphase and red lower organic layers (containing the DNA and proteins). RNA is precipitated from the aqueous layer with isopropanol. DNA is precipitated from the interphase/organic layer with ethanol. Protein is precipitated from the phenol-ethanol supernatant by isopropanol precipitation. The precipitated RNA, DNA, or protein is washed to remove impurities, and then resuspended for use in downstream applications.
RNAprotect (QIAGEN)
RNAprotect Tissue Reagent is a novel technology for immediate preservation of the gene expression pattern in animal tissues, enabling reliable gene expression analysis. After harvesting, tissues are immediately submerged in RNAprotect Tissue Reagent, which rapidly permeates the tissues to stabilize and protect cellular RNA in situ. The reagent preserves RNA for up to 1 day at 37°C, 7 days at 15–25°C, or 4 weeks at 2–8°C, allowing transportation, storage, and shipping of samples without ice or dry ice. Alternatively, the samples can be archived at −30 to −15°C or −90 to −65°C. During storage or transport in RNAprotect Tissue Reagent, even at elevated temperatures (e.g., room temperature or 37°C), the cellular RNA remains intact and undegraded. RNAprotect technology allows large numbers of samples to be easily processed and replaces inconvenient, dangerous, and equipment-intensive methods, such as snap-freezing of samples in liquid nitrogen, storage at −90 to −65°C, cutting and weighing on dry ice, or immediate processing of harvested samples.
Materials
Micropipettors set
Sterile micropipettor tips
1.7 mL Eppendorf tubes
Tubes rack
Vortex
Microcentrifuge
Cell disruptor (or tube shaking adapter for vortex)
ZymoResearch Quick RNA Fungal/Bacterial MiniPrep https://www.zymoresearch.com/products/quick-rna-fungal-bacterial-miniprep-kit?srsltid=AfmBOooGgFrroRS_vD07A_DFTK6zSHsPLI9Esj7Ajrf8aqYl-6gFlo6F
Safety concerns
This DNA extraction uses chemicals, biological materials, and equipment that pose some potential risks:
- Handle the ethanol and the kit reagents with gloves, at all times.
- Use biosafety protocols to handle the bacteria samples to prevent accidental exposure, and dispose of all the waste in a biohazard container.
- Make sure you balance the microcentrifuges correctly before you use them.
- Avoid eating or drinking, and manage spills and emergencies following the directions of the instructor.
Protocol
Samples collection and RNA extraction protocol
- Collect one tube of cells from regolith exposure experiment and centrifuge at 5000 for 5 min. Transfer 700 uL of the supernatant, without disturbing the solid phase, into an Eppendorf tube and centrifuge at 10000 rpm for 5 min. Remove the supernatant and add 300 uL of DNA/RNA Shield solution. Vortex until the cell pellet is completely resuspended.
- Add 500 uL of RNA lysis buffer and mix well.
- Transfer the mixture into a ZR BashingBead Lysis tube and treat on a bead beater for 10 min at maximum speed.
- Centrifuge the tube for 1 minute to pellet debris.
- Transfer 400 uL of the clear supernatant into a Zymo-Spin IIICG column in a collection tube and centrifuge at 10000 rpm for 1 min. Discard the column and save the flow-through.
- To the flow-through, add 400 uL of 95% ethanol and mix well.
- Transfer the mixture into a IICR column in a collection tube and centrifuge at 10000 rpm for 1 min. Discard the flow through.
- Add 400 uL of RNA Prep Buffer to the column and centrifuge. Discard the flow-through.
- Add 700 uL of RNA Wash Buffer to the column and centrifuge. Discard the flow-through.
- Add 400 uL of RNA Wash Buffer to the column and centrifuge for 2 minutes to completely remove the buffer.
- Transfer the column to an Eppendorf tube.
- Add 50 uL of elution buffer directly onto the column matrix, incubate at room temperature 1 min and centrifuge at 10000 rpm for 1 min.
- Measure the RNA concentration and run a 1% bleach agarose gel to confirm the integrity of your RNA. As a reference, undegraded RNA should show as two intense bands at approximately 2100 and 1400 bp, corresponding with the 16S and 23S subunits of the rRNA, as well as a small, diffuse band at 25 bp that corresponds to mRNA and small RNAs:
Key Takeaways
-
RNases are ubiquitous (in your gloves, in the lab surfaces, in the reagents) and degrade RNA easily. Using RNase-free reagents, tubes, and wearing gloves throughout the procedure protects RNA integrity.
-
Using reagents like DNA/RNA shield (other options are RNAprotect or flash-freezing samples in liquid nitrogen) prevents RNA degradation before extraction begins.
-
The integrity and purity of extracted RNA—checked via gel electrophoresis and spectrophotometry—determine the success of techniques like RT-qPCR and RNA-seq analysis.
Lecture slides
Download the RNAextraction_2025 to follow the pre-lab lecture.
Lab report
Download the RNAextraction_Labreport report file and submit on Canvas, following the directions of your instructor.
